Western Blot Optimization
Enhance detection and quantification of low abundance protein
The technique of western blotting illuminates molecular events including protein expression, protein localisation, protein-protein interactions or post translational modifications (PTMs). These molecular events are often very subtle. For example within an individual signaling pathway only a certain percentage of the molecules may have undergone a PTM. As such detection of proteins with low expression levels is a common problem for researchers. To help you optimize both the detection and quantification of such proteins and overall quality of the final western blot image, we have put together useful tips for the following steps:
Sample Preparation
Initial improvements in the detection and quantification of low abundant proteins can be made at the sample preparation stage. There are several steps for maximising the recovery of your target protein from the sample.
Lysis buffers
Lysis is the process in which the cell membrane is broken and the contents of the cell are re-suspended in a soluble form. The primary active agent in lysis buffer is the detergent which can be ionic or non-ionic. The best known of the ionic detergents is sodium dodecyl sulfate (SDS) while non-ionic detergents include Tween, Triton X and NP-40. Ionic detergents are considered harsher than the non-ionic detergents because of their ability to interfere with protein structure. As such their use with antibodies raised against denatured forms or synthetic peptides of the target protein is often recommended. If further denaturing is required a reducing agent (DTT or Beta mercaptoethanol) may be used to disrupt S-S bonds. Non-ionic detergents induce less protein denaturation so if their use is often recommended with primary antibodies raised against native proteins. Use of the correct lysis buffer will improve the detection of the low abundant target protein.
Table 1. Lysis Buffers
Lysis Buffer |
Detergent |
Detergent Type |
Cellular Compartment |
Antibody Immunogen |
|---|---|---|---|---|
|
RIPA |
SDS |
Ionic |
Cytoplasmic, membrane or nuclear |
Denatured protein or peptide |
|
NP-40 |
NP-40 |
Non-ionic |
Cytoplasmic |
Intact protein |
|
Tris-Trition |
Triton |
Non-ionic |
Cytoskeleton |
Intact protein |
When lysing the cell or tissue, use as little lysis buffer as possible in order to concentrate samples as much as possible. This will allow the addition of increased amounts of protein per well furthering chances of detecting the low abundant protein. However, please note that using an insufficient amount of lysis buffer may lead to an incomplete lysis of the sample. Protein recovery can vary according to the cell type and recovery method. HeLa cells contain approximately 300 pg per cell. Typical loading amounts of protein added to each well range from 10 µg (~330,000 cells) to 50 µg (~1,650,000 cells).
Table 2. Approximate Protein Recovery (based on HeLa cells, the number of cells will vary according to cell type).
Plates |
Approx. Number of Cells at Confluency |
Lysis Buffer Volume |
Approx. Protein Recovery (µg Protein) |
|---|---|---|---|
|
6-well |
1,200,000 |
200-400 |
360 |
|
12-well |
400,000 |
100-200 |
120 |
|
24-well |
200,000 |
50-100 |
60 |
|
Flasks |
|
|
|
|
T-25 |
2,800,000 |
500 |
840 |
|
T-75 |
8,400,000 |
1ml |
2,520 |
|
T-160 |
18,400,000 |
2.5 ml |
5,570 |
Determining concentration of recovered protein
There are three primary methods for determining the concentration of recovered protein. They are all colorimetric assays and can utilize known concentrations of Bovine Serum Albumin (BSA) to construct a standard curve against which samples can be evaluated.
Protein Assay |
Method |
Range |
|---|---|---|
|
Lowry |
Peptide bonds react with copper under alkaline conditions to produce Cu+ |
10-1,000 µg/ml |
|
Bradford |
Under acidic conditions Coomassie Brilliant Blue G-250 binding to protein |
1-20 µg |
|
BCA |
Reduce Cu2+ to Cu+ which binds to protein |
-0.5 µg/ml to 1.5 mg/ml |
Recovery of protein from individual cellular compartments
If the target protein is primarily expressed in a single cellular compartment (membrane or nucleus) then recovery of the proteins from the individual component will increase the chances of quantifiable detection. For recovery of nuclear proteins, Bio-Rad offers the ReadyPrep™ Protein Extraction Kit (Cytoplasmic/Nuclear). An anti-lamin A/C antibody (VMA00200) can be used as a marker to differentiate nuclear from cytoplasmic samples. In addition to a nuclear protein extraction Bio-Rad has protein extraction kits for membrane bound proteins as well as cytosolic signaling proteins.
Protein precipitation
Another strategy for increasing specific protein detection is to add a compound such as acetone that causes protein to precipitate. After centrifugation of the precipitated protein to a pellet, the supernatant is removed and the protein pellet is re-dissolved in sample buffer. One disadvantage of protein precipitation is that proteins might denature, making the pellet difficult to re-solubilize.
Gel Electrophoresis/Transfer/Blocking
Knowing the size of the target protein will indicate the gel that should be chosen and the conditions for transfer. In general Bio-Rad recommends its Criterion™ Tris-tricine precast gels for small proteins and its Criterion XT Tris-acetate precast gels for separating higher molecular weight proteins (see section below for more details).
Gel electrophoresis
Selecting the correct gel for the target protein can be a crucial step in optimizing detection and quantification. If the target protein is of low molecular weight a high percentage gel should be chosen in order to maximize protein resolution. Conversely if the target protein has a high molecular weight a low percentage gel offers the best resolution. Figure 1 lists the migration patterns of protein standards for the Criterion range of pre-cast gels.
Precision Plus Protein Unstained Standards

Fig.1. Migration patterns of molecular weight markers on Criterion Tris-tricine gels.
Examining the migration pattern of molecular weight markers enables selection of the correct gel for maximum resolution of the target protein.
Transfer
The size of the target protein should also influence the choice of the transfer protocol employed. Smaller molecular weight proteins transfer quicker than larger ones. If your target protein is on the larger end of the scale (>70 kDa) extend the transfer for as long as recommended. However do not allow the transfer process to proceed for too long or protein will be lost. In addition to the transfer time the type of membrane should also be considered. There are two types of membrane: PVDF and Nitrocellulose. In the case of detecting low abundant proteins the recommended choice is PVDF due to its greater binding capacity.
Blocking/Washing
Blocking of the membrane is required to eliminate large amounts of non-specific binding that would occur in its absence. BSA and re-suspended skimmed milk powder are commonly used to block non-specific binding on the membrane. The blocking agents are usually suspended in Tris buffered saline with Tween 20 detergent. In order to detect a low abundant protein the concentration of the blocking agent (BSA or milk) used may need to be reduced. Blocking is often performed with BSA at concentrations ranging from 1-5%. In the case of low abundant proteins this concentration can be reduced. In addition reducing the number of washes or the Tween content of the wash may also increase the signal.
Primary Antibody
An optimal concentration of primary antibody will give the highest signal within the linear range from binding of the target protein while minimizing non-specific binding. Primary antibody titration is the process of determining the optimal concentration of antibody required to detect the target protein. While most antibody suppliers recommend a particular dilution of antibody this often varies depending on the sample to be analyzed. Therefore it is advisable that sample specific primary antibody titration is performed.
Initial optimization/Antibody titration
If samples and/or antibody are in short supply dot blots (crude versions of western blots) are a useful first step in determining optimal antibody concentration range. The target protein or cell lysate mixture is added directly onto the surface of the Nitrocellulose or PVDF membrane. Protein solutions can be applied directly in a small volume. Each dot blot would contain known amounts of target protein or cell lysate. Dilution series for protein, primary and secondary antibodies should ideally be performed.
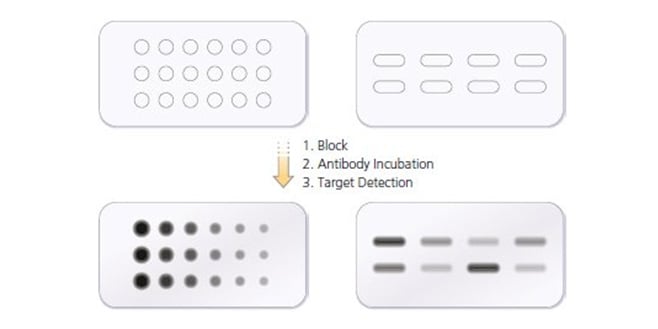
Fig. 1. A dot blot showing titration of primary antibody, secondary antibody and lysate concentration.
Once dry, dot blots are subjected to the same immuno-detection steps used for western blotting, i.e. blocking, antibody incubation, and target detection with substrate. Grey and black spots on the figure above indicate which samples are positive for the target protein and correspond roughly to the bands produced on a western blot. Selecting the dot with the highest signal gives a starting point to perform further titrations experiments around this concentration. However these methods provide no information about the size of the protein the antibody is recognizing.
Final optimization/Antibody titration
Once the initial titration is complete and a generally specific signal has been obtained the next step would be to examine the performance of the antibody in full size gels. Following the transfer, cut the membrane to generate 4 individual blots as shown below. Including a pre-stained molecular weight marker in each piece of the membrane is crucial to ensuring the signal is detected at the correct molecular weight.
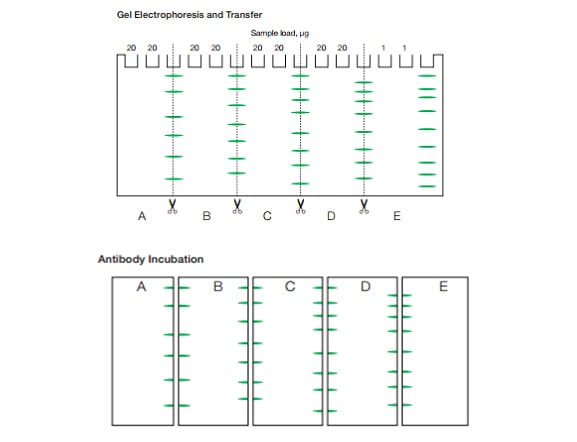
Fig. 2. Cutting the membrane for antibody titration.
Individual blots can then be incubated with varying concentrations of the primary antibody e.g. A: 1/500 B: 1/1,000 C: 1/1,500 D: 1/2,000 E: secondary only. Using a polyclonal secondary antibody can be advantageous since potential binding of several secondary antibodies to epitopes on the primary antibody provides sample amplification. Also use of a HRP labeled secondary antibody is recommended for detection of low abundant proteins as chemiluminescence is more sensitive than fluorescence. Typical dilutions of the secondary antibody range from 1/2,000 to 1/10,000 but this must be optimized through trial and error in line with the titration of the primary antibody.
In order to optimize protein quantification, we recommend using Bio-Rad’s Criterion stain free gels. The reason for this is that many house-keeping proteins (HKPs) are present at very high levels within samples. This may make their detection and that of your poorly expressed target within the linear range extremely difficult. With Criterion stain free, a total protein normalization represents a more consistent normalization method leaving you free to do long exposures to detect your weakly expressed target protein. Click here for more information on Bio-Rad stain free technology.
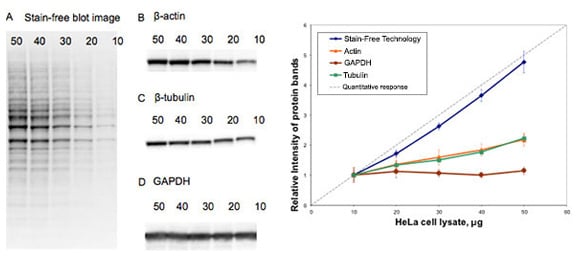
Fig. 3. Comparison of total protein normalization using Bio-Rad stain free and normalization using some common house-keeping proteins.
Signal Detection
Ultimately the goal is the detection of the antibody/antigen signal within the linear range. The linear dynamic range is the range in which signal intensity is proportional to the protein quantity on the blot.
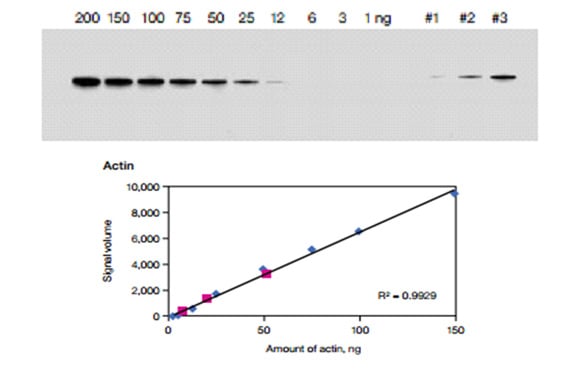
Fig. 4. Chemiluminescent blot of dilution series of purified bovine actin and three unknown amounts of actin. The background adjusted signal volume of each band in the dilution series was plotted against amount of protein to generate a standard curve. A linear fit to the data points was calculated and the corresponding R2 value for actin is shown. The high R2 value indicates a strong linear relationship between signal volume and amount of protein over a wide dynamic range.
The linear relationship between protein and signal enables quantitation with a degree of confidence. Similarly, when working with cell or tissue lysate, dilution series with varying amounts of sample can be used to ensure that quantitative measurements are conducted under linear conditions. When detecting and quantifying low abundant proteins Bio-Rad recommends customers develop the blots using Bio-Rad’s Clarity Western ECL Substrate which has long signal duration (24 hrs), a very high signal-to-noise ratio and a mid-femtogram-level sensitivity. If higher sensitivity is required, please use Bio-Rad’s Clarity Max Western ECL Substrate™ which has a low-femtogram-level sensitivity and is specifically designed for detection of very low amounts of the target protein.
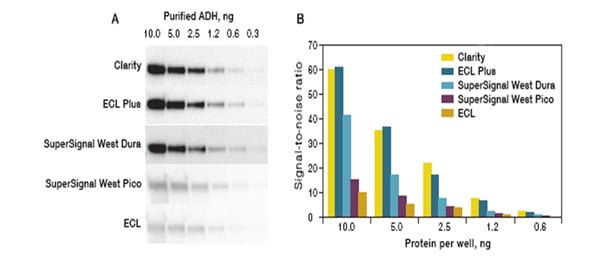
Fig. 5. Graph shows the signal detected from western blot using different ECL substrates.
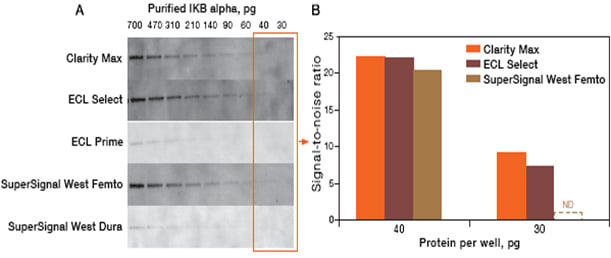
Fig. 6. Graph shows the signal detected from western blot using different high sensitivity ECL substrates.
Develop the blot using a CCD camera-based imager such Bio-Rad’s ChemiDoc™, rather than film, as a CCD camera based imager is more sensitive and detects more accurately within the dynamic linear range.

Fig. 7. Western blot showing specific and quantifiable expression.
Sample preparation, gel electrophoresis, transfer and immunoblotting must all be optimized for appropriate detection of proteins by western blot. This is even more relevant when the target protein is expressed at low levels within your sample.
Further Reading
After following the process of optimization, it should be possible to detect your antibody/antigen signal within a quantifiable linear range. For further strategies or troubleshooting western blot visit Bio-Rad's western blot resources:





