Antibody Selection and Preparation for Circulating Tumor Cell (CTC) and Rare Cell Isolation
Antibody Selection and Preparation for Circulating Tumor Cell (CTC) and Rare Cell Isolation Using the Celselect Slides Enumeration Assay
 Antibody Selection and Preparation for Circulating Tumor Cell (CTC) and Rare Cell Isolation Using the Celselect Slides Enumeration Assay
Antibody Selection and Preparation for Circulating Tumor Cell (CTC) and Rare Cell Isolation Using the Celselect Slides Enumeration Assay
The Genesis System and Celselect Slides™ utilize microfluidics and individual microchambers to efficiently capture rare cells based on their size.
After capture, enriched cells can be recovered for downstream analysis or stained on-slide for immunofluorescent applications such as enumeration and identification of various CTCs and other rare cell types using microscopy.
A critical step for successful enumeration is antibody panel design. The process involves the selection of suitable antibodies and subsequent determination of the optimal antibody concentration to use.
This guide takes you through key considerations for creating an optimized staining panel, which is part of the sample preparation step of the Celselect Slides Enumeration Direct or Indirect Stain Kits workflow (Figure 1).
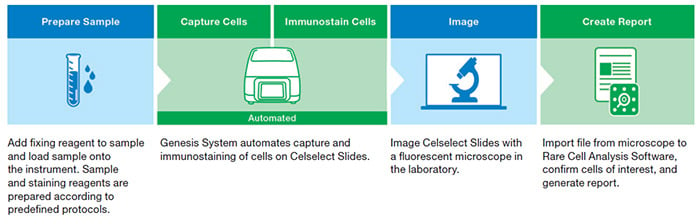
Fig. 1. Celselect Slides Enumeration Direct or Indirect Stain Kits workflow.
Considerations for Antibody Selection and Preparation
1. Target Cell
First, clearly establish what type of cell is being enumerated. There are two main cell types to choose from: epithelial and mesenchymal. Common examples of epithelial cell tumors include breast, colon, and prostate cancers. Mesenchymal cells are commonly seen in sarcomas.
- Example: MCF-7 cells are breast cancer cells, epithelial in origin, derived from adenocarcinoma of the mammary glands
2. Target Antigens
To identify your target cell type, you need to determine which expressed antigens will enable positive identification. Antigens can be expressed on the target cell surface or intracellularly, but you should also consider which antigens are not expressed by your target cell, as often a negative result can also be used to confirm identity. It is important to note that antigens are not mutually exclusive to one specific cell type. Therefore, careful analysis of antigen expression of the cell types present in your sample will be needed to ensure that the targets chosen allow detection of the target cells only.
Bio-Rad lists the immunogen on the datasheets for each antibody, and provides a handy reference guide for human immune cell marker expression. Immunogens are antigens that evoke immune responses. Knowing the immunogen will allow for appropriate antibody selection and therefore target cell identification.
- Example: for MCF-7 cells, the antigen target is cytokeratin (CK)
3. Antibody Staining Method
There are two options (Figure 2); in both cases, the antibody binding to its target antigen is visualized by a fluorescent detection system, where the fluorophore used will fluoresce at a specific wavelength (nm) when exposed to a specific light source.
- Direct — this method utilizes a fluorescently conjugated primary antibody to bind to the target antigen
- Indirect — this method uses an unconjugated primary antibody that binds to the target antigen, after which a fluorescently conjugated secondary antibody binds to the primary antibody
As more than one fluorescently conjugated secondary antibody can bind to a single primary antibody, the fluorescent signal can be significantly amplified compared to the direct staining method.
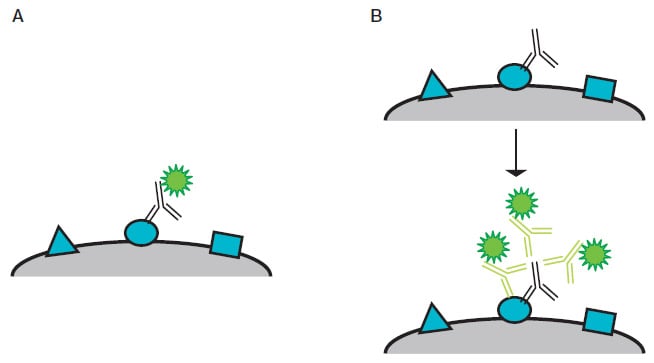
Fig. 2. Diagram of direct and indirect staining. A, direct staining — a fluorescently conjugated primary antibody binds directly to its target antigen. B, indirect staining — three fluorescently conjugated secondary antibodies indirectly detect a target antigen by binding a primary antibody bound to the antigen.
4. Primary Antibody Selection
Bio-Rad recommends that monoclonal antibodies be used for Celselect Slides Assays, as they are highly specific for an individual region of the target antigen. This is not the case with polyclonal antibodies, which detect multiple epitopes (which may lead to cross-reactivity to those same epitopes expressed on related proteins), resulting in higher background signal. Primary antibody selection depends on which staining method is used:
- Direct staining — the primary antibody must be specific for the target species (human cells) and antigen. The conjugated format of the primary antibody will need to be carefully selected; refer to consideration 6, where fluorophore selection is discussed in detail
-
Indirect staining — similarly, the primary antibody must be specific for the target species (human cells) and antigen. It is important to note that when using two or more primary antibodies in one indirect staining panel, the isotypes and/or host species should be unique to prevent cross-reactivity of the secondary antibody
- It is possible to use the same host species for multiple antibodies if a different subisotype is available. Isotypes are generalized into categories such as IgG, IgM, etc. If using the same species, each antibody should have a sub isotype of IgG1 or IgG2b
An example panel is Bio-Rad’s Celselect Slides Enumeration Indirect Stain Kit (CEL80111), which uses Mouse Anti-Human CD45 (MCA87) with an IgG2a isotype in tandem with a Mouse Anti-Human PanCK (MCA6434GA) IgG1 isotype. Here, an anti-pan-IgG secondary antibody should not be used, as cross-reactivity would occur.
5. Secondary Antibody Selection (Indirect staining only)
The isotype of the primary antibody needs to be the target antigen for the secondary antibody. For example, Bio-Rad’s CD45 antibody (MCA87) has an isotype of IgG2a; therefore, the secondary antibody must detect the IgG2a (Goat Anti-Mouse IgG2a [STAR133] would be a suitable secondary antibody in this case). The conjugated format of the secondary antibody will need to be carefully selected. Refer to consideration 6, where fluorophore selection is discussed in detail.
6. Fluorophore Selection
For successful target cell visualization, the fluorophore needs to be selected carefully.
- Understand the configuration of the fluorescent microscope being used, in order to match the fluorophores you choose to the filters available to detect them
- Select bright fluorophores that are highly photostable and have a low susceptibility to photobleaching. Conventional fluorophores such as fluorescein isothiocyanate (FITC) and R-phycoerythrin (RPE) are especially prone to photobleaching, which results in quick fading of the fluorescent signal. To reduce photobleaching, use fluorophores with high photostability
- When more than one secondary antibody is being used, it is important that the fluorophores selected fluoresce at different wavelengths so that each cell population can be identified individually. If the emission spectra of two fluorophores overlap, one fluorophore might spill/bleed into another fluorophore filter. Bleed-through can make it difficult or sometimes impossible to distinguish the two fluorophores. When selecting fluorophores, the emission and excitation spectra should therefore be checked for spectral overlap with the help of a fluorescence spectra viewer. Ideally, there should be no overlap between the fluorophore used in your immunofluorescence (IF) experiment. Bio-Rad has a tool that shows the emission spectra of fluorophores: Fluoresence Spectraviewer.
- After selecting the fluorophore for your experiment, you must decide which antigen to detect with which fluorophore. The brightest fluorophore should be reserved for the detection of the antigen with the least abundant expression level. The dimmest fluorophore should be used for detecting the most abundant antigen
7. Optimal Primary and Secondary Antibody Concentration
The optimal concentration of the antibody (primary and, if relevant, secondary antibody) will need to be determined to avoid high levels of nonspecific (off-target) background staining from excess antibody or loss of signal intensity from too little antibody, both of which can prevent visualization of your target cell. Each antibody will need to be titrated to determine the appropriate concentration for target cell identification.
Antibody Titration and Experiment Setup
As a starting point for the titration, the recommended dilution for immunofluorescence stated on the product datasheet provided by the antibody supplier can be used. If a recommended dilution is not available, the starting dilution for the titration of the Celselect Slides Assay can be determined by performing a titration in an IF experiment. During the process of determining the optimum antibody concentration and experimental setup, you may need up to six Celselect Slides plus a further two Celselect Slides to run the assay.
Key Points
- Only one concentration of the titration can be performed at any one time. Begin with the recommended starting dilution.
- Always include positive and negative controls in every experiment. Remember that the antibodies for the negative controls will need to be titrated.
- When determining the optimum antibody concentration, run the experiment with a representative cell line that is known to express the target antigen.
- Repeat with an antibody at a different dilution if the signal is too high or too low.
- Run the Direct or Indirect Enumeration Protocol for each antibody being tested. Ensure you follow the guidance exactly, or the results will be invalid.
Table 1 outlines an example titration and assay setup for the detection of PanCK-positive cells (expressed on CTCs) using the Celselect Indirect Enumeration Protocol.
Slide Number |
Primary Antibody |
Secondary Antibody |
Purpose |
|---|---|---|---|
|
1 |
|
Detection antibody for the PanCK antibody (FITC-conjugated IgG1 antibody [STAR132F]) |
Rule out false-positive staining by the secondary antibody alone |
|
2 |
|
Detection antibody for CD45 antibody (DyLight 650-conjugated IgG2a antibody [HCA310D650]) |
Rule out false-positive staining by the secondary antibody alone |
|
3 |
Detection antibody for
|
Rule out crosstalk between the two secondary antibodies |
|
|
4 |
Anti-PanCK antibody (MCA6434GA) |
Detection antibody for PanCK antibody (FITC-conjugated IgG1 antibody [STAR132F]) |
|
|
5 |
Anti-CD45 antibody (MCA87) |
Detection antibody for CD45 antibody (DyLight 650-conjugated IgG2a antibody [HCA310D650]) |
|
|
6 |
Detection antibody for
|
Confirm panels work on representative cell lines known to express target antigen |
FITC, fluorescein isothiocyanate; IgG, immunoglobulin G.
Note that these antibodies can be tested in IF first to determine the best primary and secondary antibody concentrations, to give an optimal starting concentration.
After each slide assay finishes, analyze the slide with a fluorescent microscope of choice. Bio-Rad recommends Agilent’s BioTek Lionheart LX Automated Microscope with BioTeK Gen5 V3.14 Software, which has protocols that can be used to scan Celselect Slides. Furthermore, Bio-Rad’s Rare Cell Analysis Software is compatible with the scanned images from this system.
After obtaining the images, Rare Cell Analysis Software can be used to determine target cell counts. To use this software, ensure all image files are one of the following file types: TIFF, JPEG, PNG, or BMP.
If background fluorescence is observed for an antibody (Figure 3), the gain or integration time may be too high.
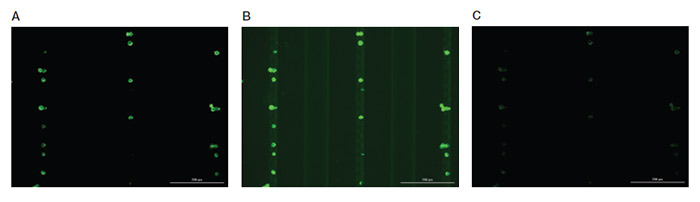
Fig. 3. Representative images of MCF-7 cells stained with anti-PanCK antibody and acquired at different parameters. A, optimized scanning parameters showing clear staining pattern of MCF-7 cells with balanced brightness and contrast. B, high level of background signal due increased exposure time, integration time, and camera gain. C, image acquired using low exposure time, resulting in dim fluorescence signal that limits visualization of cells of interest. Scale bar = 200 μm.
To reduce the background, try lowering the gain and integration time parameters and rescanning before adjusting the antibody concentration. If lowering these parameters does not eliminate the background fluorescence, titrate the antibody down to a lower concentration and then retry scanning.
If no background fluorescence is noted for each single antibody tested, run the panel of antibodies in duplicate against the cell line of interest in whole blood, as per the example assay (Table 2).
Table 2. Final assay confirmation setup.
Slide Number |
Primary Antibody |
Secondary Antibody |
Purpose |
|---|---|---|---|
|
7 |
Detection antibody for
|
Confirm panels work on the cell line of interest (MCF-7 cells) |
|
|
8 |
Detection antibody for
|
Replicate slide to double-confirm that the panels work on the cell line of interest (MCF-7 cells) |
FITC, fluorescein isothiocyanate; IgG, immunoglobulin G.
Once the slides have been analyzed on a fluorescent microscope, determine the target cell counts for each slide. If they are comparable, the antibody panel has been optimized and can be used for further testing on this sample and target cell type, using these antibodies at the optimized concentration. It should be noted that new cell and sample types (even if also epithelial in origin) will need separate antibody optimization.
Figure 4 shows the ideal antibody staining result using Bio-Rad’s Standard Indirect Enumeration Panel:
- Nuclear Marker — 4',6-diamidino-2-phenylindole (DAPI) (blue) should be present, indicating that a cell is present in the Celselect Slides chamber. Note that DAPI is already in chamber number 10 in the cartridge as part of the kit
- CTC Marker — PanCK detected by FITC should be present, indicating detection of target cell type
- White Blood Cell Marker — CD45 detected by DyLight 650 should not be present, confirming that the cell detected is not a contaminating white blood cell
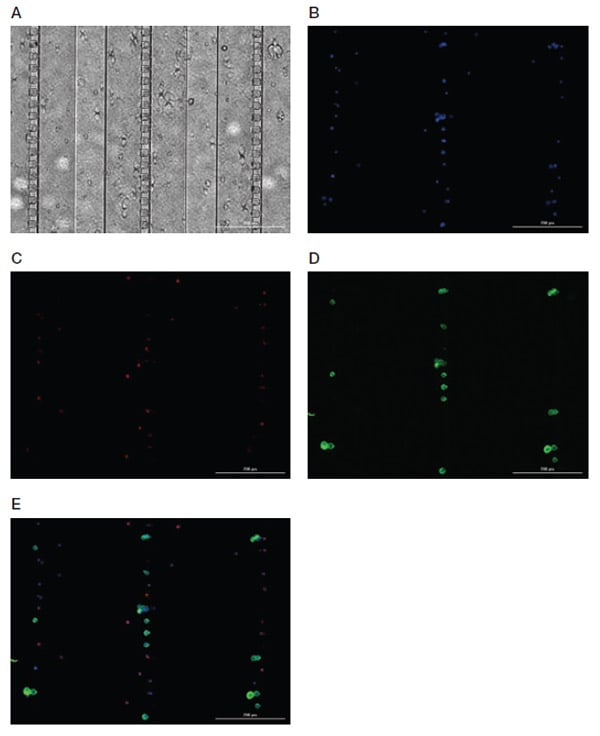
Fig. 4. Representative images of Celselect Slides CTC Staining Panel. MCF-7 target cells spiked in human whole blood are identified using CTC marker (anti-PanCK antibody) and detected by FITC-conjugated IgG1 antibody. White blood cells are identified using leukocyte marker (anti-CD45 antibody) and detected by DyLight 650–conjugated IgG2a antibody. A, bright field image; B, DAPI image; C, DyLight 650 image; D, FITC image; E, merged image. Scale bar = 200 μm. CTC, circulating tumor cell; DAPI, 4',6-diamidino-2-phenylindole.
In Summary
Bio-Rad’s Celselect Slides Enumeration Assay is an effective tool to identify target cell populations using size selection and antibody staining.
By ensuring that the antibody panel is optimized, users can be confident that they are getting highly sensitive and specific identification of the target cell population.
For further information on Bio-Rad’s Celselect Slides Enumeration Panel and Rare Cell Analysis Software, see: