Apoptosis: Late Phase of Apoptosis
Late Phase Overview
During the late phase of apoptosis, the intrinsic and extrinsic pathways merge. The late stage of apoptosis is characterized by DNA fragmentation (karyorrhexis), which starts upon completion of nuclear condensation (pyknosis). It is mediated by both caspase dependent and independent routes. In the late phase significant morphological and phenotypical changes also occur. These changes and the associated molecular events can be studied by several applications including flow cytometry, western blot and imaging techniques.
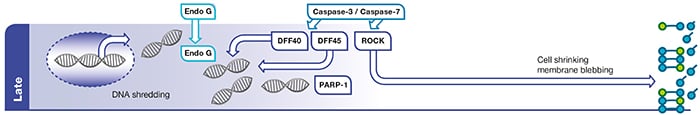
Fig. 1.
Late
phase of apoptosis. Effector caspases (caspase-3 and caspase-7) activate caspase activated DNAases (CADs) such as DNA fragmentation factor 40 (DFF40). Endonuclease G (Endo G) relocates to the nucleus and cleaves DNA. Caspase-3 cleavage of poly (ADP-ribose) polymerase-1 (PARP-1) inhibits its ability to repair DNA damage. Effector caspases also activate
Rho associated
kinase (ROCK) which leads to cell shrinkage and membrane blebbing.
Late Phase of Apoptosis
Caspase-3 translocates to the nucleus and interacts with several DNA interacting proteins such as poly (ADP-ribose) polymerase (PARP-1) and caspase activated DNAases (CADs) such as DFF40.
Under non-apoptotic conditions, the DNAases are in an inactive state. DFF40 is maintained in this inactive state by binding with the inhibitory factor DFF45. Caspase-3/caspase-7 activation releases the active DFF40 from the inhibiting factor DFF45. Caspase activated endonucleases such as DFF40, also known as CAD (caspase activated DNase), cleave or break the genomic DNA into oligonucleosome fragments or multiples of approximately 200 base pairs. When the genomic DNA from such late apoptotic cells is extracted and run on agarose gels, a “DNA ladder” pattern can be observed. DNA fragmentation can also be observed using TUNEL (Terminal deoxynucleotidyl transferase dUTP Nick End Labeling) assays.
Caspase-3 cleaves the 116 kDa isoform of PARP-1 at the DEVD site to generate 85 kDa and 24 kDa fragments preventing DNA repair. Endo G is involved in caspase-independent apoptosis via DNA degradation when released from the mitochondrion to nucleus during apoptosis.
Analysis of Late Phase Events
Poly (ADP-ribose) polymerases (PARPs) are DNA-dependent nuclear enzymes that transfer negatively charged ADP-ribose moieties from cellular nicotinamide-adenine-dinucleotide (NAD+) to a variety of protein substrates, altering protein–protein and protein-DNA interactions. The most studied of these enzymes is PARP-1, a nuclear enzyme which binds DNA and plays an important role in promoting DNA repair. PARP-1 facilitates recruitment of proteins involved in both base excisional repair (BER) and homologous recombination (HR). PARP-1 is involved in the base excision repair pathway by catalyzing the synthesis of poly (ADP-ribose) and modifying nuclear proteins. Inhibition of DNA repair enzymes by caspases occurs alongside caspase mediated DNA fragmentation. Caspase-3 cleavage of PARP inhibits its ability to repair DNA damage.
The enzyme which fragments DNA during apoptosis is the caspase activated DNAases (CAD) also known as DNA fragmentation factor subunit beta (DFFB/DFF40). DFF is a heterodimeric protein of 40 kDa (DFFB) and 45-kD (DFFA) subunits. DFFA is the substrate for caspase-3 and triggers DNA fragmentation during apoptosis. DFF becomes activated when DFFA is cleaved by caspase-3. The cleaved fragments of DFFA dissociate from DFFB, the active component of DFF. DFFB has been found to trigger both DNA fragmentation and chromatin condensation during apoptosis. Alternatively spliced transcript variants encoding distinct isoforms have been found for DFFB but the biological validity of some of these variants has not been determined.
Endo G is a mitochondrial specific nuclease that relocates to the nucleus during apoptosis where it cleaves chromatin and DNA.
Viability Dyes in Apoptosis
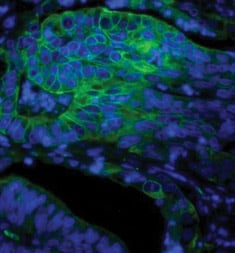
Fig. 2. Paraffin section of human colon adenocarcinoma stained with Mouse Anti-Human Cytokeratin 18, detected with Goat Anti-Mouse IgG:A488 and counterstained with PureBluTM DAPI.
Viability dyes are often used in combination with apoptotic assays to discriminate cells undergoing apoptosis and cells which have already compromised membranes. Two examples of such dyes are Propidium Iodide (PI) and 7-aminoactinomycin (7-AAD), which can be used in flow cytometry and are often combined with Annexin V Apoptosis Assays to distinguish dead cells from those undergoing apoptosis. To visualise apoptosis in microscopy, DAPI can be used alongside other apoptotic markers such as mitochondrial membrane potential dyes.
DAPI is a dye that can be used to visualize nuclear changes and assess apoptosis. It binds strongly and selectively to the minor groove of adenine-thymine regions of DNA. When bound to double-stranded DNA, DAPI absorbs light at 359 nm (ultraviolet light) and emits at 461 nm (Figure 2). Bound DAPI has a fluorescence intensity approximately 20 fold higher than that of unbound DAPI. As the apoptotic cell membrane is compromised, more DAPI enters the cell and stains a stronger blue color. The differing nuclear morphology of apoptotic cells such as chromosome condensation and fragmentation also helps with visual identification of apoptotic cells stained with DAPI.
PI is a fluorescent dye that binds to DNA and RNA in a stoichiometric ratio. PI is not membrane permeable and therefore requires the compromised membranes of dead cells to bind to DNA. Upon excitation with an appropriate light source, dead cells exhibit higher fluorescence intensity than live cells, thereby enabling clear discrimination of these populations by flow cytometry. Dead cells can then be excluded from further analysis and also sorted from cell populations of interest. The spectral properties (excitation 535 nm/emission 617 nm) enable use in both single color and multicolor flow experiments.
7-AAD is a fluorescent dye that has a high affinity for GC-rich regions of double stranded DNA. 7-AAD is not membrane permeable and therefore requires the compromised membranes of dead cells to bind to DNA. Upon excitation with an appropriate light source, dead cells exhibit higher fluorescence intensity than live cells, thereby enabling clear discrimination of these populations by flow cytometry.
For a complete listing of Bio-Rad viability dyes, visit the cell viability page.



